Biological Structure and Imaging
Research summary
Cryo-EM is inaugurating a new era for structural biology through its abilities to reveal atomic-resolution structures of biological samples without crystallization. The increasing demand for general applicability and higher throughput at the atomic-resolution level poses challenges for cryo-EM computing technologies. High-dimensional parameter estimation is still a challenging but key factor in obtaining reliable results. Failures in reconstruction or classification, as well as problems such as low resolution and over-refinement, are usually caused by bad parameter estimation and are recognized as an issue in 3D-reconstruction algorithms.
We are focusing on the following research directions:
A Particle-filter framework for robust cryo-EM 3D reconstruction
Single particle reconstruction algorithm design
Cryo-ET reconstruction and subtomogram averaging
Incremental learning-based protein particle picking algorithm
3D-reconstruction algorithms for cryo-electron tomography
EPicker, a particle picking software based on neural networks
Representative works
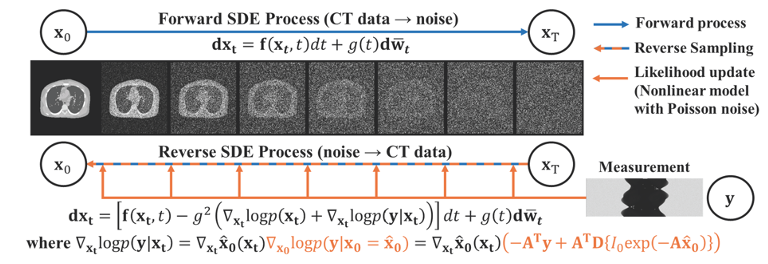 |
We propose a plug-and-play diffusion posterior sampling method that integrates a general nonlinear measurement CT imaging model. With combination of trained prior and likelihood function to arrive at a posterior score function that can be used to sample the reverse-time diffusion process.
|
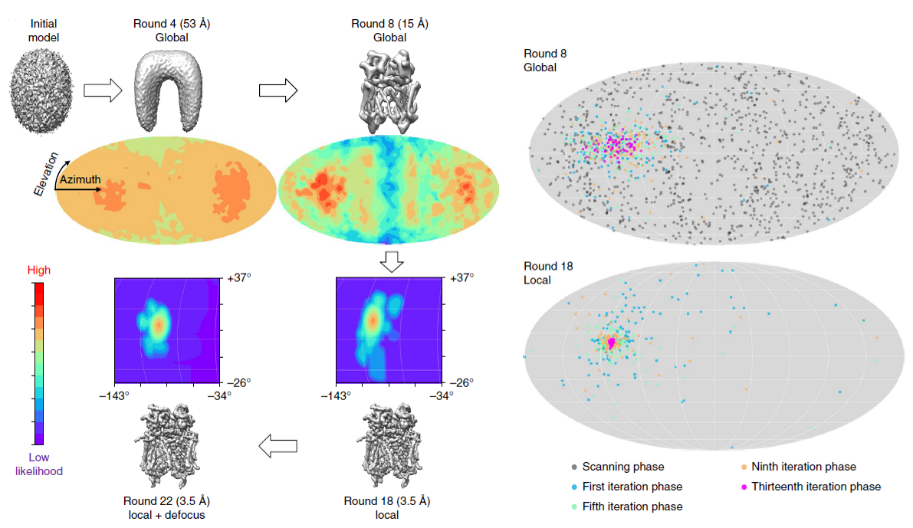 |
We introduce a particle-filter algorithm for cryo-EM, which provides high-dimensional parameter estimation through a posterior probability density function (PDF) of the parameters given in the model and the experimental image.
|
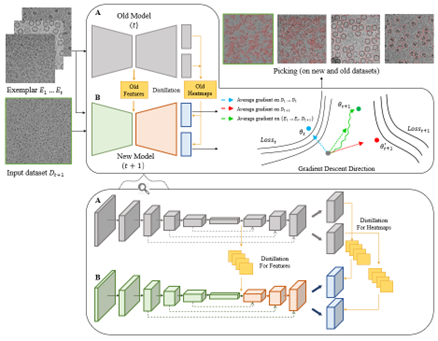 |
We developed a particle picking software based on CenterNet. This program can generate samples for continual learning with the aid of 2D classification for further discrimination and reconstruction. The optimized and highly automated pipeline is able to recognize particles, vesicles and filaments in single particle micrographs.
|
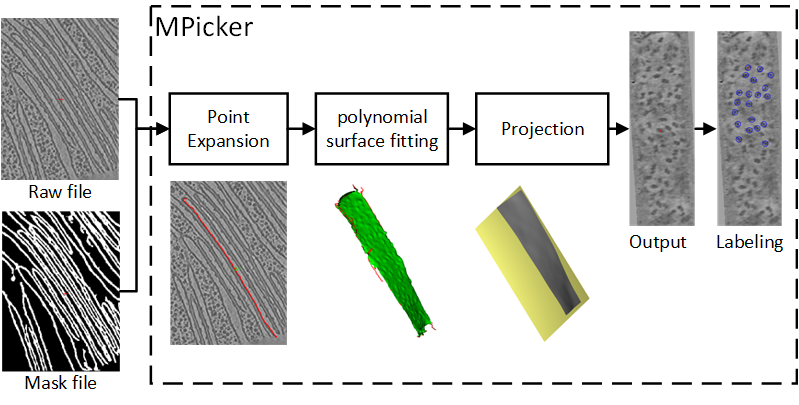 |
We developed a membrane-flattening method and the corresponding software, MPicker, designed for the visualization, localization, and orientation determination of membrane proteins in cryoET tomograms. In MPicker, we integrated approaches for automated particle picking and coarse alignment of membrane proteins for sub-tomogram averaging.
|
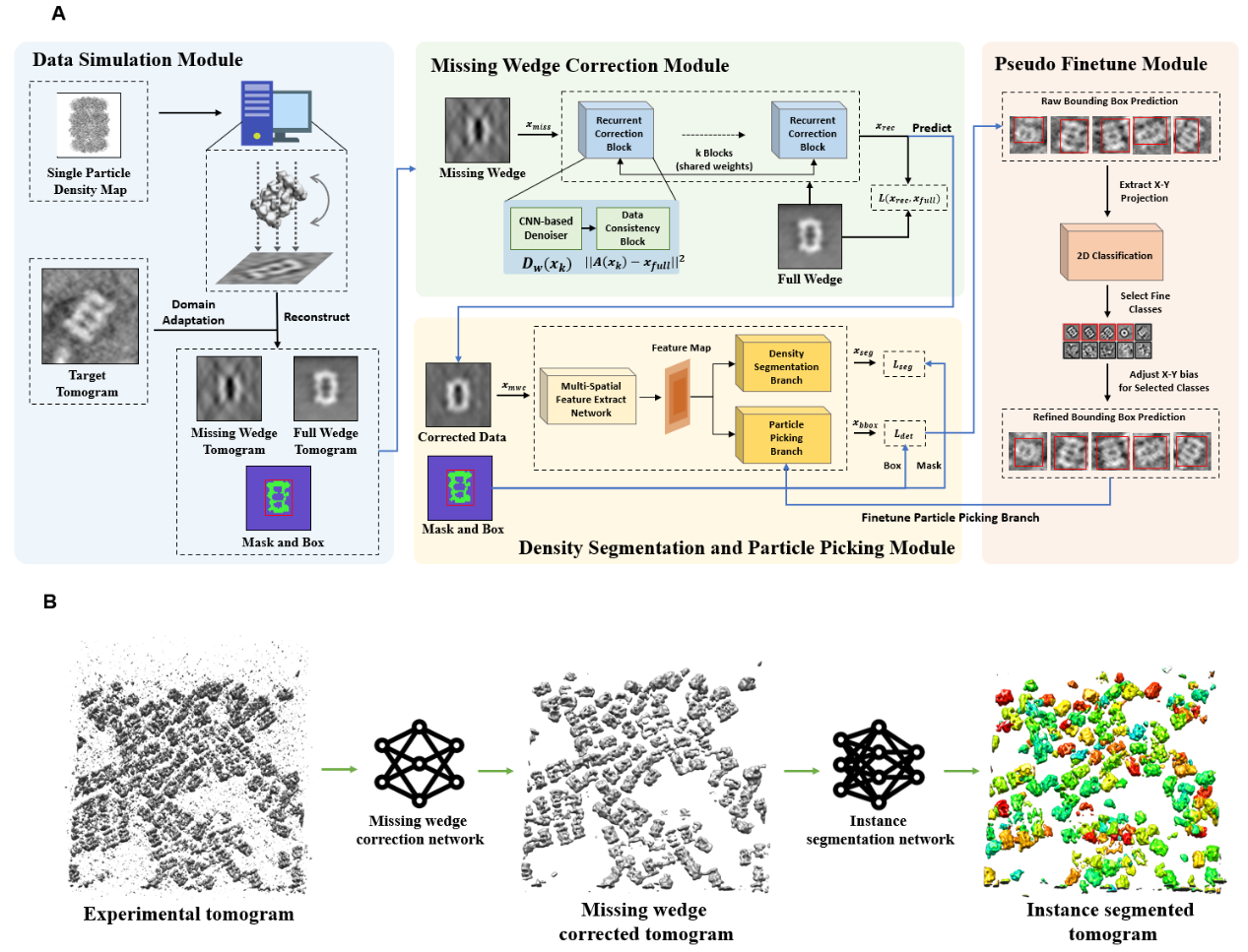 |
We propose a simulation data-based framework for in-situ particle picking in reconstructed tomography. The pipeline can generate simulation datasets according to the given particle map for training a missing wedge correction and an instance segmentation network, which can be applied to recognize the locations of the target particles with high accuracy in the reconstructed tomography.
|
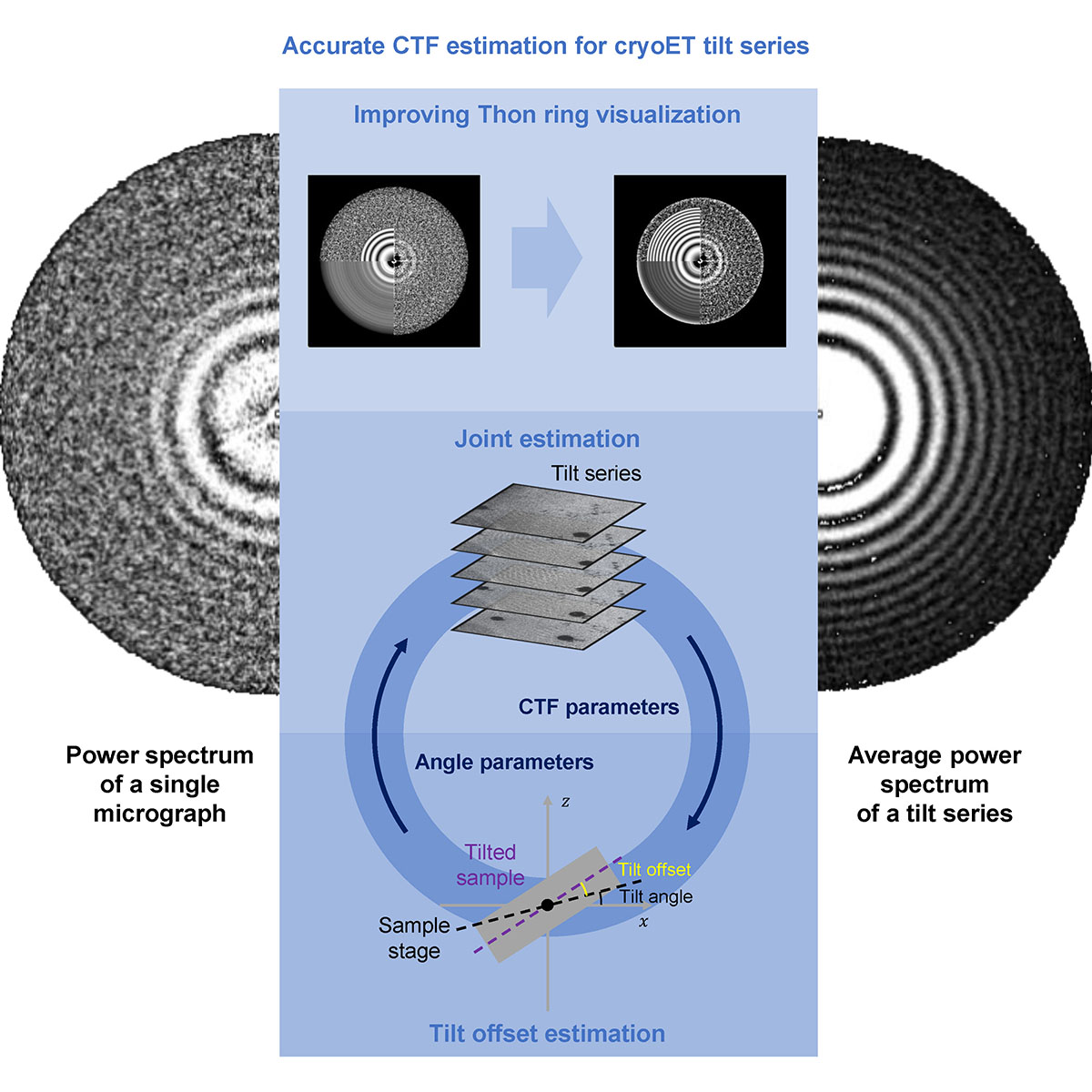 |
We propose a tilt-series-based joint CTF estimation method implemented in the new software CTFMeasure. The joint estimation method combines all Thon-ring signals in a tilt series to improve the estimation accuracy. By using an objective function involving the CTF parameters and geometric parameters of a cryoET tilt series, CTFMeasure can estimate the CTF parameters of each micrograph and the absolute tilt angle offset of the lamellar sample relative to the sample stage plane, which is usually the glancing angle used during focused ion beam (FIB) milling.
|