Single Prticle Reconstruction for Cryo-EM
Research summary
Cryo-EM is inaugurating a new era for structural biology through its abilities to reveal atomic-resolution structures of biological samples without crystallization. The increasing demand for general applicability and higher throughput at the atomic-resolution level poses challenges for cryo-EM computing technologies. High-dimensional parameter estimation is still a challenging but key factor in obtaining reliable results. Failures in reconstruction or classification, as well as problems such as low resolution and over-refinement, are usually caused by bad parameter estimation and are recognized as an issue in 3D-reconstruction algorithms.
We are focusing on the following research directions:
A Particle-filter framework for robust cryo-EM 3D reconstruction
Single particle reconstruction algorithm design
Cryo-ET reconstruction and subtomogram averaging
Incremental learning-based protein particle picking algorithm
3D-reconstruction algorithms for cryo-electron tomography
EPicker, a particle picking software based on neural networks
Representative works
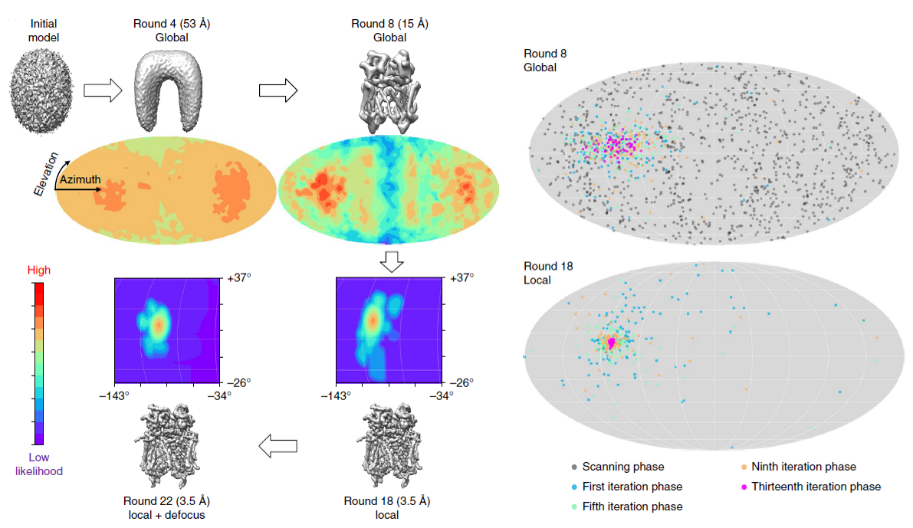 |
We introduce a particle-filter algorithm for cryo-EM, which provides high-dimensional parameter estimation through a posterior probability density function (PDF) of the parameters given in the model and the experimental image.
|
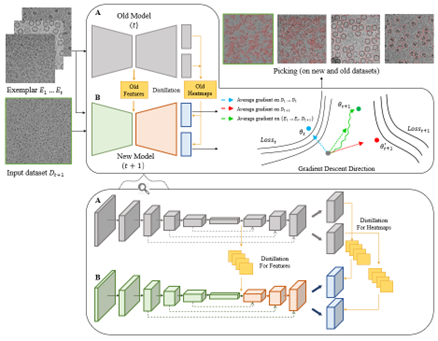 |
We developed a particle picking software based on CenterNet. This program can generate samples for continual learning with the aid of 2D classification for further discrimination and reconstruction. The optimized and highly automated pipeline is able to recognize particles, vesicles and filaments in single particle micrographs.
|
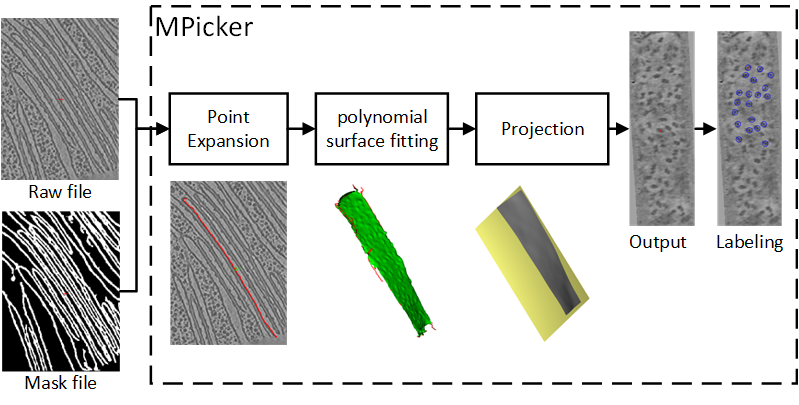 |
We develop a software called MPicker for the visualization of membrane proteins based on point expansion and polynomial surface fitting methods. This software can extract membranes with different structure and visualize the membrane proteins that are difficult to perceive in general circumstance. |
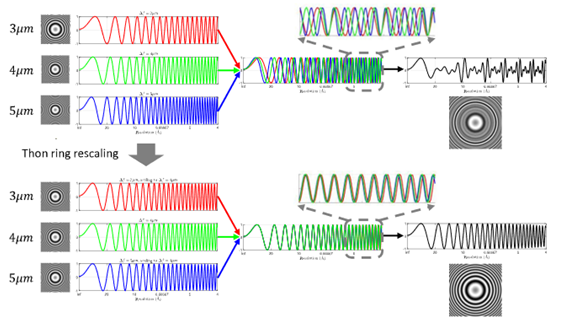 |
We propose a new CTF estimation software for cryo-ET tilt-series. Tilt-series-based joint estimation with Thon ring rescaling technique is applied to alleviate aliasing in high spatial frequency and recover more Thon rings, which leads to more accurate estimation of CTF parameters. The resolution of the protein structure after subtomogram averaging improves by 20% without further CTF refinement. |
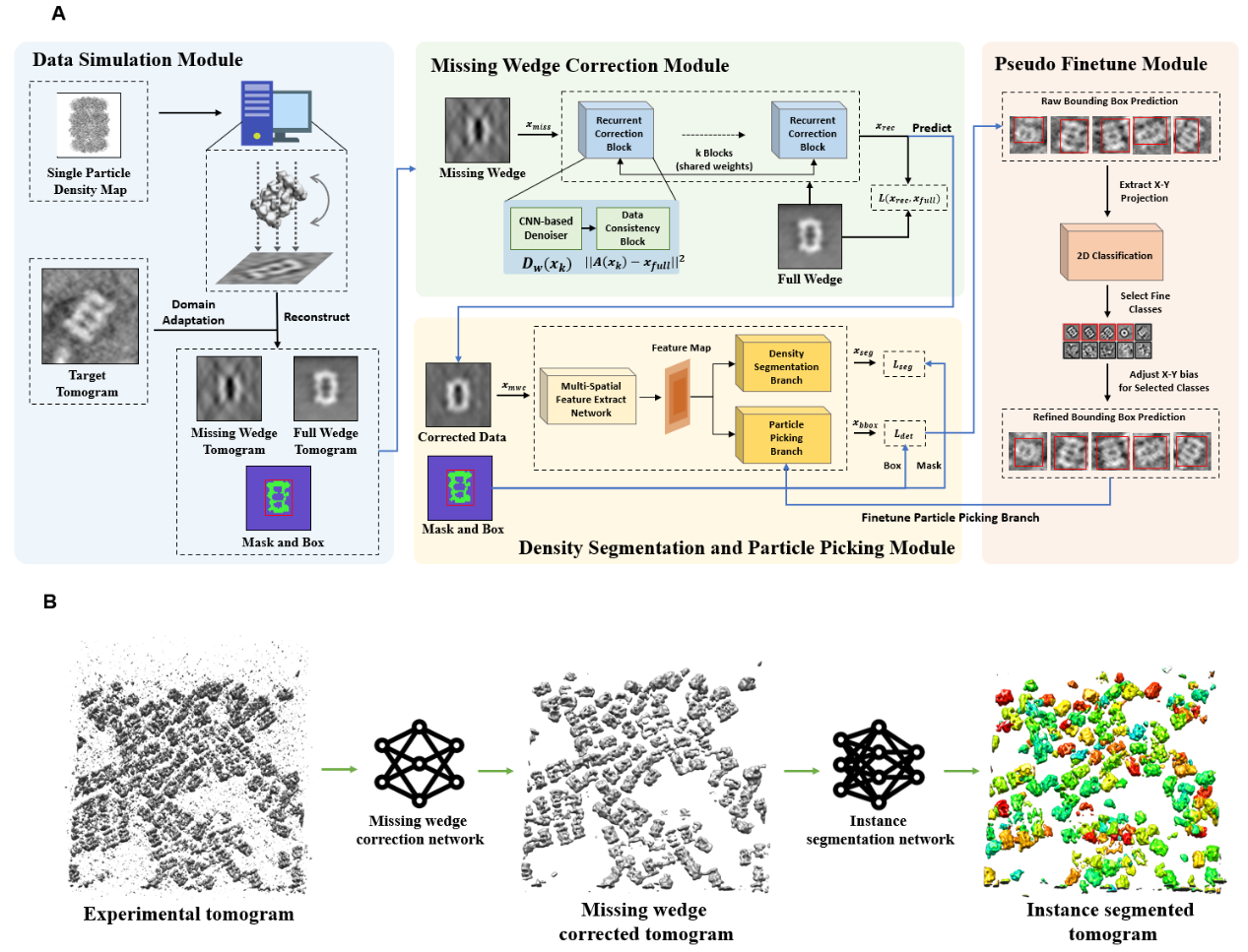 |
We propose a simulation data-based framework for in-situ particle picking in reconstructed tomography. The pipeline can generate simulation datasets according to the given particle map for training a missing wedge correction and an instance segmentation network, which can be applied to recognize the locations of the target particles with high accuracy in the reconstructed tomography. |
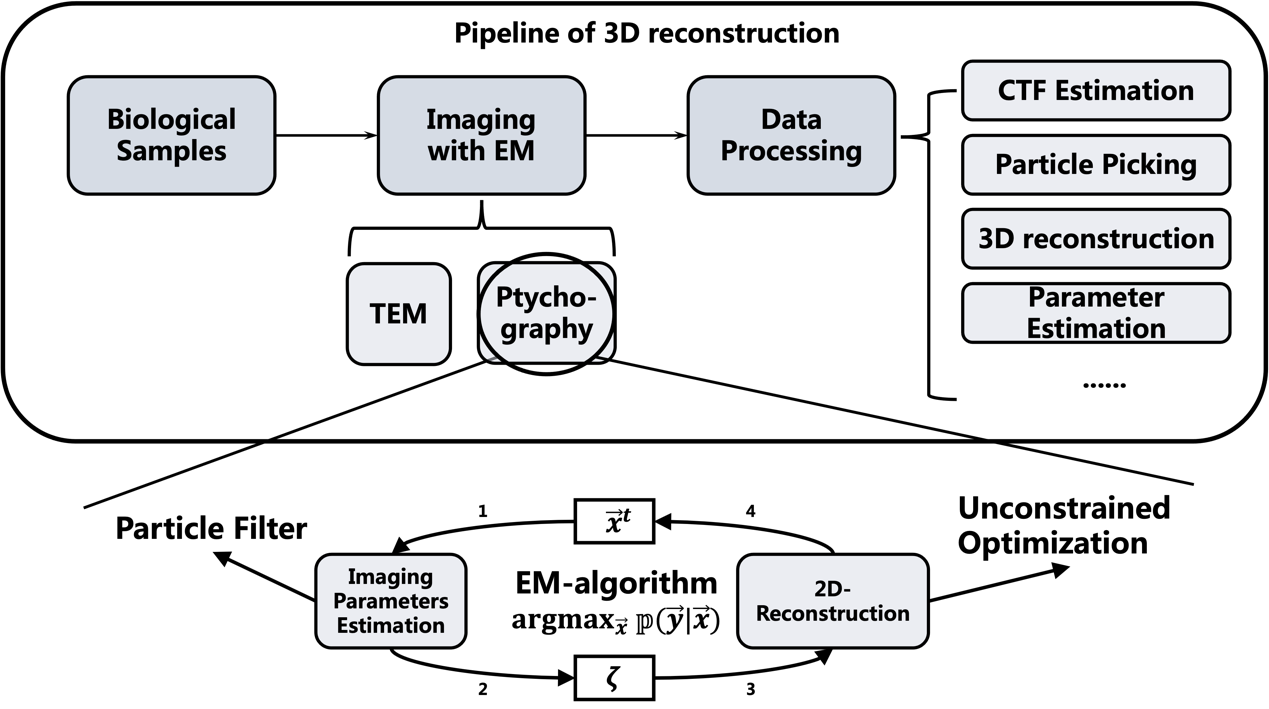 |
We introduce EM-algorithm into ptychography, which provides more accurate estimation on imaging parameters in simulation datasets. With this technology, we focus on ptychographic imaging on the biological samples, which may enable 3D-reconstruction to reach higher resolution than that of 3D-reconstruction with TEM images. |